Nodular goiter may increase the risk of thyroid cancer, but the genetic factors contributing to nodular goiter are not well understood. There is an overexpression of H19 lncRNA in goiter tissue and its target remains unknown. In this study, we attempted to identify a new target for H19 in the context of goiter development.
MethodsUsing interaction energy calculations, the interaction between NKX2-1 mRNA and H19 lncRNA was examined. Putative microRNAs were found at the H19 lncRNA target site with the highest affinity for NKX2-1. RNAseq data was analyzed to determine the tissue specificity of gene expression. Samples were taken from 18 goiter and 18 normal tissues during thyroidectomy. The expression of NKX2-1 was determined by RT-qPCR using specific primers.
ResultsThe interaction between NKX2-1 and H19 was characterized by six local base-pairing connections, with a maximum energy of −20.56 kcal/moL. Specifically, the sequence that displayed the highest affinity for binding with H19 overlapped with the binding site of has-miR-1827 to NKX2-1. It was found that NKX2-1 is exclusively co-expressed with H19 in normal thyroid tissue. As compared to adjacent normal tissues, nodular goiter tissues have a significant overexpression of NKX2-1 (relative expression = 1.195, p = 0.038).
ConclusionNKX2-1 has been identified as the putative target of H19 lncRNA, which is overexpressed in nodular goiter tissues significantly.
Level of Evidence4.
Goiter is a potential cause of thyroidectomy, and its molecular and genetic foundations remain unknown. In addition, 15.6% of patients undergoing thyroid surgery are later diagnosed with thyroid cancer, a rate that is higher than expected.1 The research pertaining to thyroid diseases has predominantly concentrated on well-differentiated and undifferentiated thyroid cancers,2,3 thereby neglecting the benign nodular goiters.
The gene NK2 Homeobox-1 (NKX2-1, ENSG00000136352) encodes a transcription factor that expresses in the early stages of tissue development, such as the lung, forebrain, hypothalamus, and primarily the thyroid gland.4 Homozygous animals lacking NKX2-1 exhibited absence of the thyroid gland during birth and perished shortly thereafter.5 In siblings with congenital hypothyroid goiter, NKX2-1 expression is diminished, and the gene is associated with congenital hypothyroidism in newborns.6–9 When considering cancer, the NKX2-1 gene exhibits two distinct and opposing impacts. Firstly, it hinders cell proliferation and mobility.10 Conversely, overexpression of this gene is also responsible for lung adenocarcinoma progression.11 Therefore, NKX2-1 plays an important role in the progression of multiple diseases, but its influence on benign nodular goiters is not fully understood.
A recent study in Brazil utilizing RNA-seq analysis on thyroid tissue afflicted with multinodular goiter indicated that 70 genes demonstrated differential expression.12 Out of these genes, 61 were down-regulated while the remaining nine were up-regulated. A particular gene, H19 long non-coding RNA (H19 lncRNA, ENSG00000130600), appears to be overexpressed exclusively in nodular goiter tissues. According to the expression atlas (https://www.ebi.ac.uk/gxa/home), NKX2-1 and H19 are highly co-expressed in normal thyroid tissue (expression level = 219 TPM and 95 TPM, respectively). The lung is the only other tissue that co-expresses the two genes, but at much lower levels (expression levels = 44 TPM and 5 TPM, respectively). Moreover, in the thyroid tissues catalogued by the expression atlas, NKX2-1 expression surpasses that of H19. This suggests that the gene could be targeted downstream by lncRNA. On the other hands, our unpublished data revealed that seven MicroRNAs (miRs) have putative binding sites at the 3'UTR of the NKX2-1 gene. Among these miRs tested was hsa-miR-1827 (ENSG00000221476), whose overexpression has been associated with the progression and metastasis of breast, lung, and liver cancer.13–15 A particularly attractive theme is the possibility of hsa-miR-1827 sponging with H19 lncRNA. Based on the findings of the expression atlas and the Brazilian study, we hypothesized that NKX2-1 might be a plausible target for H19 lncRNA and it must be also over-expressed in goiter tissues.
Here we provided novel insights into the potential connection between NKX2-1 mRNA and H19 lncRNA. We also assessed NKX2-1 expression in benign goiter and adjacent normal tissues. We identified six potential binding sites at the 3'UTR of NKX2-1 gene for H19 binding. One of these sites, which had the highest affinity for H19, overlapped with the binding site of Has-miR-1827. Moreover, it was observed that the expression of NKX2-1 was notably elevated in nodular goiter tissues when compared to adjacent normal tissues.
MethodsPrediction of mRNA-lncRNA interactionsThe RIblast service offered by the lncRRsearch web server was implemented to forecast complex RNA-RNA interactions through the utilization of stabilizer energy obtained from the hybridization of two RNAs.16 A specified threshold interaction energy of −16 kcal/moL was applied. The visualization of each local base-pairing interaction was accomplished through VARNA.17 The H19 binding site with the highest affinity for NKX2-1 (and the lowest binding energy) was used for miRNA prediction.
Identification of miRNA with the same binding site as H19 against NKX2-1MicroRNA target prediction database (miRDB) was used to predict putative miRNAs targeting NKX2-1.18 The “custom prediction” option of miRDB was used to identify potential miRNAs that target the H19 binding site with the highest affinity for the 3’UTR of NKX2-1.
Tissue-specific expression analysisIn a cohort of 960 individuals, the RNAseq data underwent analysis through the Genotype-Tissue Expression (GTEx) portal.19 This analysis aimed to estimate the expression levels of human H19 lncRNA and NKX2-1 mRNA across 54 distinct tissues. To calculate the median expression of the genes, the GTEx_Analysis_2017-06-05_v8_RNASeQCv1.1.9_gene_reads.gct.gz file was used (https://www.gtexportal.org/home/downloads/adult-gtex/bulk_tissue_expression).
Tissue samplesAt Al-Zahra and Sina hospitals in Isfahan, each patient who received a recommendation for a thyroidectomy was mandated to sign a consent form. The Helsinki protocol was followed (ethical code IR.UI.REC.1398.058). In this study, goiter and normal thyroid tissue samples were excluded based on a checklist of criteria. The indicators investigated included the presence of excessive blood in the tissue, fat surrounding the tissue, and the very small dimensions of the excised tissue. An expert pathologist endorsed the inclusion of both goiter and normal thyroid tissue samples, which had been examined on two distinct microscopic slides. Histopathological examinations of 36 samples of thyroid tissue revealed 18 to be benign goiter, while the remaining 18 were classified as normal thyroid. Initially, there were many more participants, but they were later excluded due to dissatisfaction. The histopathological analysis was performed in a third-party laboratory. According to the 7th edition of the American Joint Committee on Cancer Tumor-Node-Metastasis (TNM) staging system, tumor staging was acknowledged.
RNA extraction and cDNA synthesisFresh tissues were immersed in RNAlater fixation solution (Ambion Life Science Company, USA) according to the manufacturer’s instructions, and incubated at 4 °C for 24 h. For long-term preservation, tissues were stored at −70 °C once they had been removed from the fixation solution. After treatment with RNAlater, tissues weighing 50 ± 5 mg were rinsed in a standard saline solution (0.85% NaCl). Following the immersion in liquid nitrogen, the tissues were pulverized in a mortar. Total RNA was extracted using Biobasic Company’s extraction solution (Bio Basic-Canada, Lot: BS410A-N116DR0J). To eliminate residual genomic DNA, DNase treatment was carried out according to the manufacturer’s instructions (Thermo Fisher Scientific, Germany, Lot: 00645766). cDNA was synthesized using random hexamers as per the manufacturer’s instructions (Thermo Fisher Scientific, USA, Lot: 00645766).
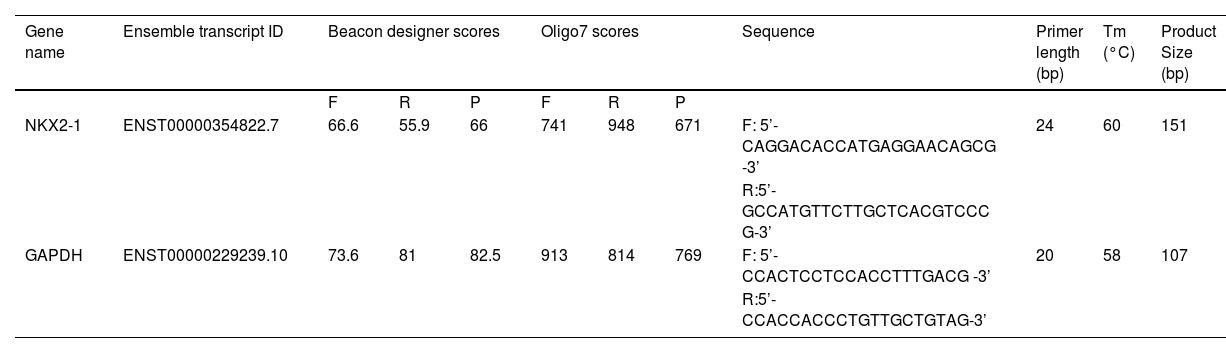
Primer designingThe exon-junction primers were created using Beacon Designer 8 software (Premier Biosoft USA). In order to ensure the absence of primer dimers and additional loop formation, Oligo7 software (Molecular Biology Insights, USA) was employed to select primers devoid of secondary structures. Table 1 provides a comprehensive explanation of the primers utilized in the investigation. To ensure data uniformity, the GAPDH gene was selected as a calibrator. The primer melting temperature was determined through temperature gradient PCR.
The characteristics of the primers.
| Gene name | Ensemble transcript ID | Beacon designer scores | Oligo7 scores | Sequence | Primer length (bp) | Tm (°C) | Product Size (bp) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| F | R | P | F | R | P | ||||||
| NKX2-1 | ENST00000354822.7 | 66.6 | 55.9 | 66 | 741 | 948 | 671 | F: 5’- CAGGACACCATGAGGAACAGCG -3’ | 24 | 60 | 151 |
| R:5’- GCCATGTTCTTGCTCACGTCCC G-3’ | |||||||||||
| GAPDH | ENST00000229239.10 | 73.6 | 81 | 82.5 | 913 | 814 | 769 | F: 5’- CCACTCCTCCACCTTTGACG -3’ | 20 | 58 | 107 |
| R:5’- CCACCACCCTGTTGCTGTAG-3’ | |||||||||||
A list of genes and the accession numbers of their corresponding transcripts is provided. Beacon designer and oligo software scores of primers are added. The length of the primers, melting temperature and size of the PCR product are included.
F, Forward primer; R, Reverse primer; P, PCR Product.
RT-qPCR was conducted in a final volume of 12 µL using an Amplicon kit (SYBR Green RealQ Plus 2× Master Mix, Denmark, Lot: A323402). Initial denaturation and enzyme activation were performed at 95 °C for 15 min. The first 40 cycles consisted of primer extension at 72 °C for 30 s, annealing at Tm temperature, and denaturation at 95 °C. All reactions were performed in triplicate with a non-template negative control.
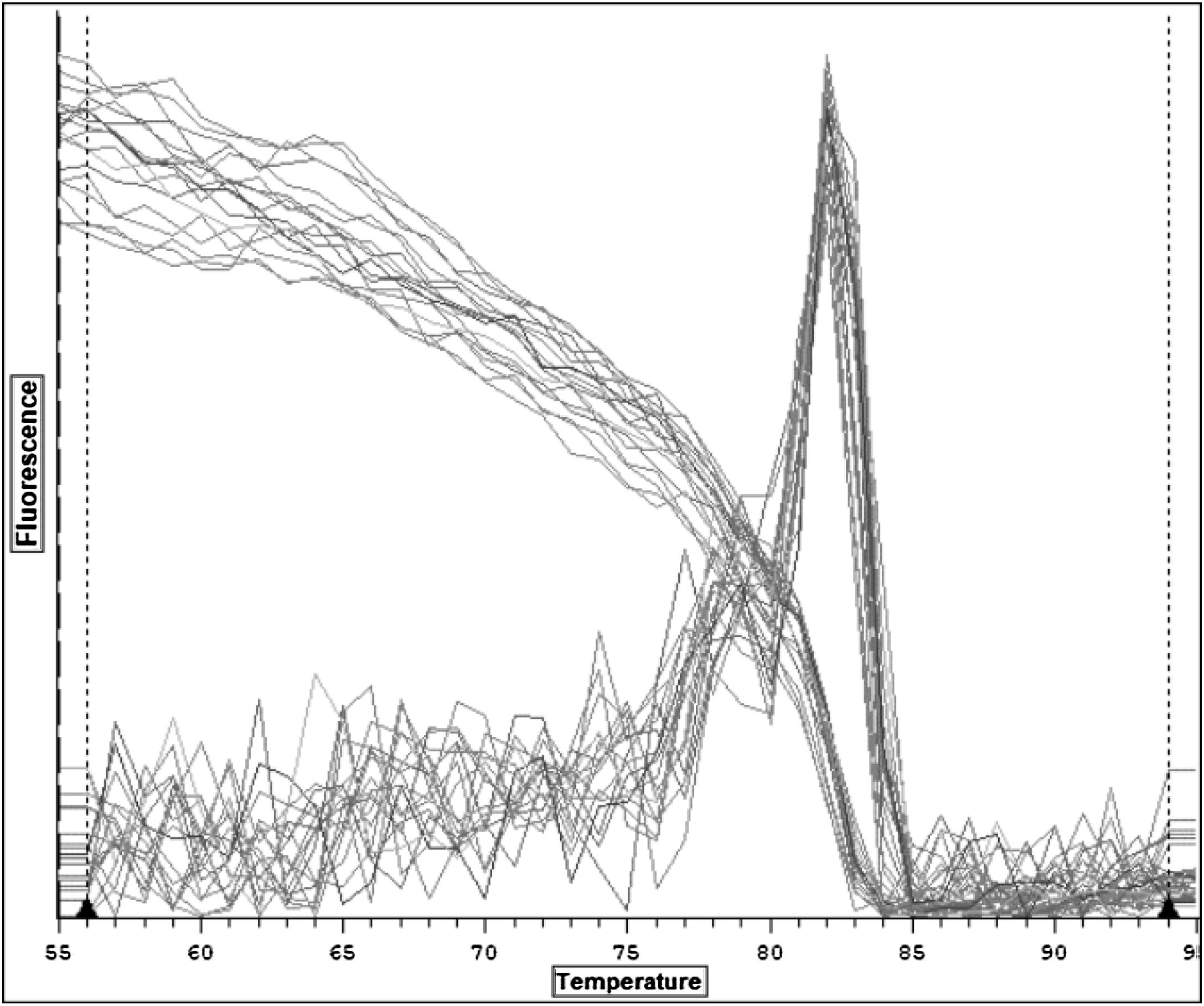
Melting curve analysisThe specificity of the RT-qPCR reaction was assessed by melting curve analysis. This commenced at 55 °C and grew by 1 °C until it reached 95 °C. A fluorescent alteration in the green channel was detected and documented for each degree Celsius that the temperature surpassed the baseline level.
Statistical analysisUsing the t-test, the mean expression level of the NKX2-1 gene in goiter tissues was compared with that in their corresponding adjacent normal tissues. The relative expression of the NKX2-1 gene was evaluated using the Livak method.20
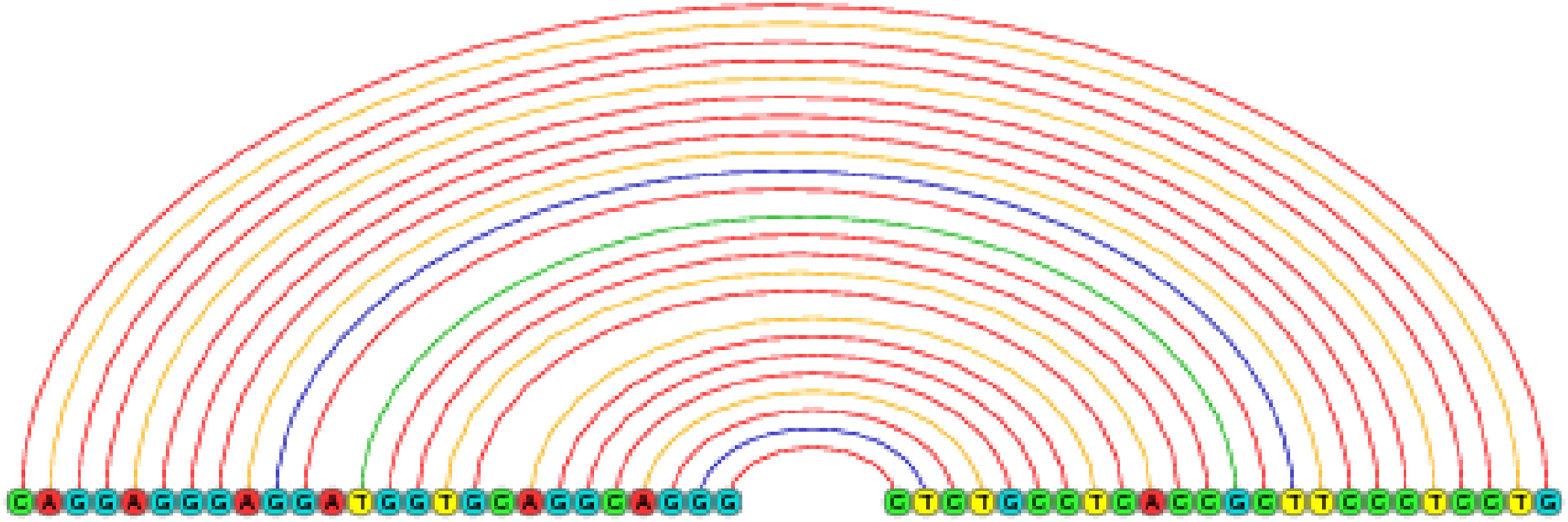
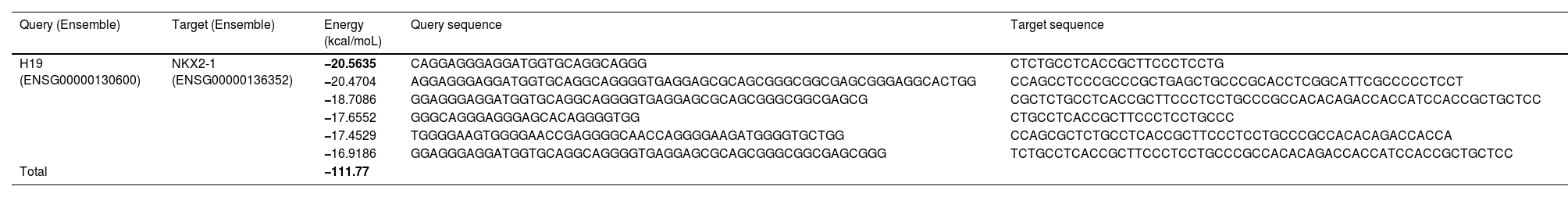
ResultsNKX2-1 interacts with H19 lncRNARIblast predicted six local base-pairing interactions between NKX2-1 and H19, as outlined in Table 2. The cumulative interaction energy of these local base-pairing was −111.77 kcal/moL. Among these interactions, the maximum local base-pairing interaction energy was −20.56 kcal/moL, which is visually depicted in Fig. 1. Additionally, supplementary Figs. S1‒S5 offer graphical representations of the remaining five local base-pairing interactions.
NKX2-1/H19 lncRNA interactions information.
| Query (Ensemble) | Target (Ensemble) | Energy (kcal/moL) | Query sequence | Target sequence |
|---|---|---|---|---|
| H19 (ENSG00000130600) | NKX2-1 (ENSG00000136352) | −20.5635 | CAGGAGGGAGGATGGTGCAGGCAGGG | CTCTGCCTCACCGCTTCCCTCCTG |
| −20.4704 | AGGAGGGAGGATGGTGCAGGCAGGGGTGAGGAGCGCAGCGGGCGGCGAGCGGGAGGCACTGG | CCAGCCTCCCGCCCGCTGAGCTGCCCGCACCTCGGCATTCGCCCCCTCCT | ||
| −18.7086 | GGAGGGAGGATGGTGCAGGCAGGGGTGAGGAGCGCAGCGGGCGGCGAGCG | CGCTCTGCCTCACCGCTTCCCTCCTGCCCGCCACACAGACCACCATCCACCGCTGCTCC | ||
| −17.6552 | GGGCAGGGAGGGAGCACAGGGGTGG | CTGCCTCACCGCTTCCCTCCTGCCC | ||
| −17.4529 | TGGGGAAGTGGGGAACCGAGGGGCAACCAGGGGAAGATGGGGTGCTGG | CCAGCGCTCTGCCTCACCGCTTCCCTCCTGCCCGCCACACAGACCACCA | ||
| −16.9186 | GGAGGGAGGATGGTGCAGGCAGGGGTGAGGAGCGCAGCGGGCGGCGAGCGGG | TCTGCCTCACCGCTTCCCTCCTGCCCGCCACACAGACCACCATCCACCGCTGCTCC | ||
| Total | −111.77 | |||
A RIblast analysis predicted six local base-pairing interactions between H19 lncRNA (query) and NKX2-1 (target). Maximum and minimum local base-pairing interaction energies are −20.56 kcal/moL and −16.92 kcal/moL, respectively. Interacting sequences of the query and target are shown.
An exemplification of the maximal local interactivity of base-pairing between NKX2-1 mRNA and H19 lncRNA is presented. The red color denotes the complementary base pairing between guanine and cytosine, while the yellow color represents that of adenine and thymine. Additionally, there exists an inadequate base pairing, denoted by green and blue colors.
We identified seven microRNA molecules targeting NKX2-1 including hsa-miR-7107-3p, hsa-miR-6753-3p, hsa-miR-323a-5p, hsa-miR-1286, hsa-miR-6797-5p, hsa-miR-1249-5p and hsa-miR-1827. Only hsa-miR-1827 showed similar binding site on the 3'UTR of NKX2-1 as H19 lncRNA (Fig. 2).
The sequence of human NK2 Homeobox-1 (NKX2-1, ENST00000354822.7, NM_001079668.3) along with its details. Translated sequences are shown in blue, flanking sequences in green, intronic sequences in gray, and Untranslated Regions (UTRs) at the 5' and 3' ends of the gene are shown in red. The sequence with yellow highlights is the main binding site for H19 lncRNA. H19 and has-miR-1827 share overlapping sequence, which is bolded and underlined.
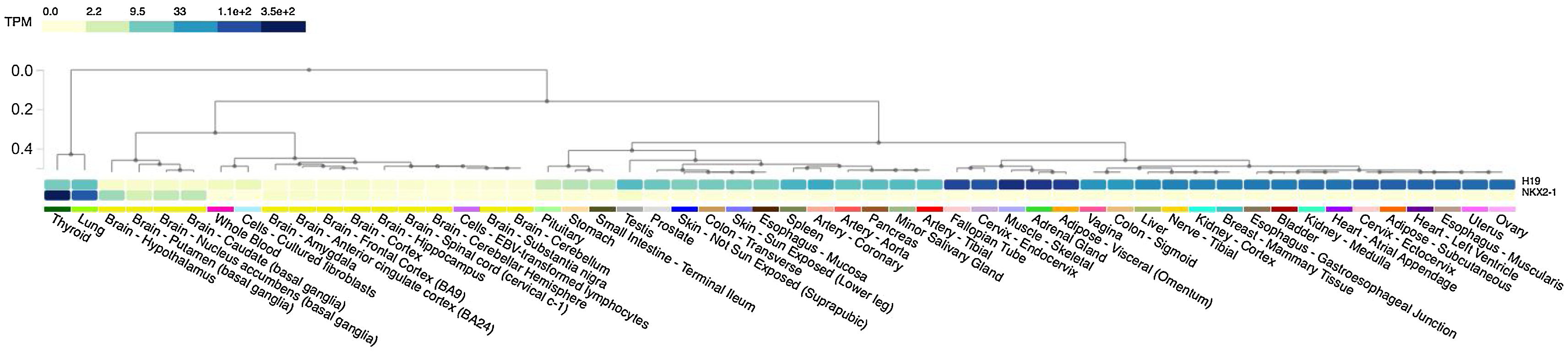
Simultaneous overexpression of both NKX2-1 and H19 was observed in normal thyroid tissue, as illustrated in Fig. 3. An analysis of tissue-specific gene expression patterns using the GTEx portal. There is a high level of expression of the transcription factor NKX2-1 in thyroid tissue (median TPM = 352.6), a lesser degree in lung tissue (median TPM = 89.67), and a very low level of expression in various parts of the brain tissue (median TPM = 5.622‒2.180). H19 lncRNA can be found in a variety of tissues, but it appears to co-express with NKX2-1 primarily in normal thyroid and lung tissues. H19 demonstrated expression in diverse tissues including skeletal muscle, adipose, testis, breast, ovary, prostate, and thyroid tissue. Equal expression of H19 is documented in normal thyroid (median TPM = 8.887) and lung (median TPM = 12.06) tissues. As a summary, NKX2-1 and H19 co-expressed in thyroid tissue, but H19 was expressed at a much lower level than NKX2-1.
An analysis of tissue-specific gene expression patterns using the GTEx portal. The transcription factor NKX2-1 has been observed to be exclusively expressed in thyroid tissue (median TPM = 352.6), to a lesser extent in lung tissue (median TPM = 89.67) and different parts of brain tissue (median TPM = 5.622–2.180). H19 lncRNA can be found in a variety of tissues. However, it appears to co-express with NKX2-1 primarily in normal thyroid tissues and then in the lungs.
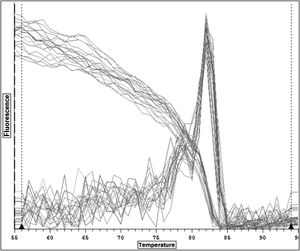
The temperature peak at 83 °C matched the bioinformatics prediction, confirming the primer specificity in RT-qPCR (Fig. 4). Notably, the absence of non-specific peaks suggests the absence of primer dimers and non-specific products.
Analysis of RT-qPCR dataThe NKX2-1 gene expression levels were evaluated in goiter tissues and their respective adjacent normal tissues, as depicted in Table 3. Our findings indicate a significant upregulation of the NKX2-1 gene in goiter tissues when compared to normal tissues, as evidenced by the relative expression value of 1.195 (p = 0.038).
Comparison of NKX2-1 gene expression between nodular goiter tissues and adjacent normal tissues.
| Gene name | Relative expression | p-Value | Result |
|---|---|---|---|
| GAPDH | 1 | ‒ | |
| NKX2-1 | 0.172 | 0.04 | Down-regulation |
As compared to adjacent normal tissues, goiter tissues exhibit a downregulation of the NKX2-1 gene.
GAPDH, Glyceraldehyde 3-Phosphate Dehydrogenase; NKX2-1, NK2 Homeobox-1.
The presence of thyroid nodules in the later stages of adulthood carries a significant risk of malignant lesions.21 However, the genetic basis of nodular goiter remains unknown. A specific mutation in NKX2-1 has been associated with both benign multinodular goiter and malignant Papillary Thyroid Cancer (PTC).22 There is a correlation between the SNP rs944289 located in the NKX2-1 gene and an increased risk of thyroid cancer.23 There has been a report of NKX2-1 gene rearrangements in T-cell acute lymphoblastic leukemia.24 These findings suggest an oncogenic role for the NKX2-1 gene. But unexpectedly, we observed that the NKX2-1 gene expression in PTC demonstrated no significant alterations (p = 0.885).25
To elucidate our observation in conjunction with the knowledge that NKX2-1 functions as a marker for thyroid differentiation, an investigation was conducted to ascertain the potential regulatory molecule(s) responsible for controlling NKX2-1 expression. In the meantime, substantial overexpression of the H19 lncRNA was reported in cases of nodular goiter,12 with the targets of this non-coding RNA remaining elusive. The findings of our investigation have validated that the H19 regulatory lncRNA engages in interactions with the 3'UTR of NKX2-1 mRNA at six distinct sequences. Remarkably, the sequence exhibiting the most pronounced affinity for H19 coincides with has-miR-1827 binding site.
Moreover, RNAseq analysis revealed that H19 is co-expressed with NKX2-1 in normal thyroid tissue. Then we found that normal thyroid tissue expresses the highest level of NKX2-1, followed by lung tissue at a quarter level. Exaggerated expression in thyroid tissue is significance since the gene induces tissue specificity via transcription and regulation of thyroid-specific genes.26 The expression of the NKX2-1 gene exhibits a significant correlation with follicular production, viability of follicular cell precursors, and the expression of thyrocyte-specific genes during maturity.27,28 It is interesting to note that mice with NKX2-1 expression loss frequently presented with adenomas, alongside thyroid follicle atrophy.29 Taking all these factors into consideration, NKX2-1 gene expression caused thyroid-specific genes to transactivate and to keep differentiation status of the tissue.
The decrease in NKX2-1 expression is linked to the transformation of thyroid adenomas into thyroid carcinomas, and the gradual decrease in its localization in the nucleus corresponds to thyroid tumor dedifferentiation.30 Moreover, a study found that NKX2-1 expression was higher in 100% of differentiated follicular adenomas, but in 0% of undifferentiated anaplastic carcinomas.31 The findings of our current investigation indicate a marked elevation in the expression level of the NKX2-1 gene in tissues exhibiting benign nodular goiter when compared to their adjacent normal tissues. We propose that the overexpression of the NKX2-1 gene may be an effective strategy for preventing tumor progression through the maintenance of the differentiation status of nodular goiter cells. As a further explanation, our differentiated malignant PTC tissues were shown to not differently expressed NKX2-1,25 and microarray meta-analysis supports the finding (adj. p value = 1). It appears that NKX2-1 is essential for maintaining the normal configuration and function of differentiated thyroids as well as inhibiting the aggressive expansion of the thyroid. Differentiated PTC tissues are cancerous; therefore NKX2-1 must be expressed at the same level as normal tissues. Nodular goiter is also differentiated as normal tissues, but is not cancerous as it overexpresses NKX2-1 to prevent tumor progression. When faced with a pre-malignant phenotype, nodular goiter cells may attempt to maintain their non-malignant status by overexpressing NKX2-1. Consequently, it is postulated that the upregulation of NKX2-1 may serve as a ultimate defensive mechanism employed by goiter cells to restrict their aggressiveness and avert the onset of cancer.
Mechanistically, it could be proposed that H19 lncRNA overexpression in nodular goiter could sponge hsa-miR-1827 and induce the expression of the thyroid differentiation marker, NKX2-1. We did not, however, approve this matter within our laboratory; nevertheless, we are strongly dedicated to implementing it in our future endeavors. According to the authors, MYCL1,32 MDM2/TP53,33 RBX1/CRKL14 and SFRP113 are the only identified target genes of hsa-miR-1827. Our research findings suggest that the maintenance of the highest level of differentiation status in the benign goiter could potentially serve as an approach employed by impoverished benign cells to impede malignancy advancement.
ConclusionWe discovered for the first time that the NKX2-1 mRNA was a target for the H19 lncRNA regulatory molecule. Moreover we found that has-miR-1827 binds to an overlapping sequence as H19 binds to NKX2-1. The expression levels of NKX2-1 mRNA were also found to be markedly higher in the tissues of nodular goiters compared to normal tissues.
Author’s contributionsS-M.J.: Conceptualization, design, data analysis, interpretation, financial support, manuscript drafting, critical revision for critical intellectual contents, and final approval of the manuscript.
FundingThis study was conducted at University of Isfahan and was supported financially by the Departments of Research, Technology and Graduate Offices (Grant number: A1097997).
Ethics approval and consent to participateAll patients recommended for thyroid surgery had to sign a consent form. The acquisition of human tissues was approved by University of Isfahan institutional review board. All the experiments and procedures performed in this study specially but not limited to human participants were in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Authors sincerely thank the volunteers for their participation.
Data availability statementBulk tissue expression data are available at GTEx portal via the following URL: https://www.gtexportal.org/home/downloads/adult-gtex.
Conflicts of interestThe author declares no conflicts of interest.
Peer Review under the responsibility of Associação Brasileira de Otorrinolaringologia e Cirurgia Cérvico-Facial.